Computational Linguistics for COVID-19 !
Table of Contents
Literature Based Discovery
People
Activities
Our goal is to enrich a given corpus of COVID-related biomedical literature with biomedical entities.
Our annotation process is based on an efficient dictionary-based lookup (OGER) combined with a deep learning approach trained on existing corpora, for example the CRAFT corpus of the University of Colorado.
Our terminologies are derived from the major life science databases using our Bio Term Hub, which allows us to maintain up-to-date dictionaries synchronized with the original resources.
Our current annotation pipeline generates annotations for several entity types:
- cell lines
- clinical drugs (RxNorm)
- cells
- molecular processes
- sequences
- organ/tissue
- chemicals
- Gene Ontology (GO)
- organisms
- proteins
Click here to access the annotated documents
One interesting observation is that there has been an "infodemic" about COVID19 not only in the media outlets, but also in the scientific literature. The graph below shows the number of COVID19-related papers published on PubMed daily from the beginning of the year. Notice that the weekly peaks and troughs are simply due to the fact that articles are not published on weekends!
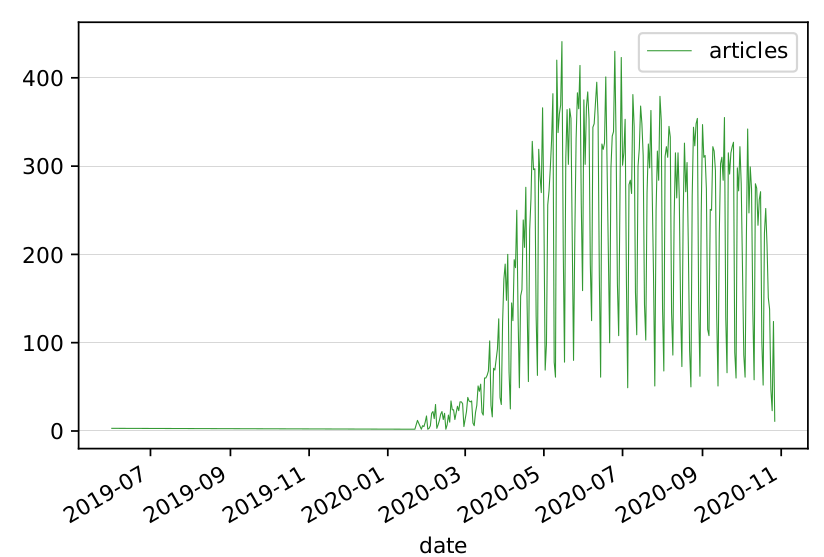
Timeline
- Update of our processed version of the LitCovid dataset. It contains about 35'313 abstracts.
- Update of our processed version of the LitCovid dataset. It contains about 25'000 abstracts.
- Datasets updated until today.
- We have updated our annotated LitCovid dataset (now containing 5630 abstracts)
- We have completed the annotation of the PMC subset of Litcovid. Find it here.
- Our online annotation platform OGER now includes COVID specific terminology. You can also use it as a web service, try it out!
Our OGER+BioBERT annotations are now accessible on a local brat installation.
See a screenshot below:

- We have submitted our (improved) OGER+BioBERT annotations of the LitCovid dataset to Europe PMC.
- We have annotated the LitCovid dataset with OGER+BioBERT and published our results on PubAnnotation, a tool developed by DBCLS, Tokyo (group Jin-Dong Kim):
Datasets
We have been working with two recently released datasets.
Our annotated datasets
- Terminology
- Our own COVID19 terminology, in BTH format, partially derived from a dataset provided by the Lab of Patrick Ruch (HES-SO, Geneva).
- Annotated corpora.
- We have processed the LitCovid corpus with our entity recognition tools. Click here for details and downloads, in different formats.
- Only a few of the abstracts contained in LitCovid have also a full text accessible from PubMed Central. We have processed this subset of full text paper (which we refer to as LitCovid/PMC). The results are available here.
Who are we?
This page is currently maintained by the NLP Group at the Dalle Molle Institute for Artificial Intelligence (IDSIA).
The work described in this page was initially carried out by the Biomedical Text Mining group at the Institute of Computational Linguistics, University of Zurich. It is now being continued at IDSIA where the PI of the group (Fabio Rinaldi) and some group members have moved.
For additional information about the tools and research activities described in this page, please contact Fabio Rinaldi.